Our hand-made FFT code from FFT-II is very fast (order ![]() for a full Fourier transform of a length –
for a full Fourier transform of a length –![]() input), but we made no optimizations. The julia FFTW library (which uses routines from the C FFTW library) is highly optimized and very fast. I wrote our code in the previous post to emulate its input and output as closely as possible so that comparison is easy.
input), but we made no optimizations. The julia FFTW library (which uses routines from the C FFTW library) is highly optimized and very fast. I wrote our code in the previous post to emulate its input and output as closely as possible so that comparison is easy.
using FFTW
using Plots
N=256
L=5
t = range(-L, stop=L, length=N)
# -5.0:0.0392156862745098:5.0
# note 10/255=0.0392156862745098
k=1:256
shiftedks = fftshift(fftfreq(N)*N)
# -128:1:127
# our input
signal = cos(2*π*3 .* t)
FFTsignal = fftshift(fft(signal))
plot(x, f, title = "signal(t)", xlabel = "Time", ylabel = "f(t)")
plot((1/(2*L))*shiftedks, abs.(FFTsignal), title = "FFT", xlabel = "ω/2π", ylabel = "F(ω)")
The function fftshift(fftfreq(N)*N) creates a set of symmetric (about ![]() ) frequencies, which we did by hand, based on the size of the sample
) frequencies, which we did by hand, based on the size of the sample ![]() which again must be a power of
which again must be a power of ![]() . We do some rescaling of the shifted k’s when we make the plots of the FFT’ed signal, which again is two nice delta functions.
. We do some rescaling of the shifted k’s when we make the plots of the FFT’ed signal, which again is two nice delta functions.
Reading/parsing FITS files
Many of you will need to analyze spectra stored in FITS file formats. You can get a nice FITS file viewer “fv” that reads headers and prints out datafields, or you can use the FITSIO julia library to read out useful things in the header.
I downloaded a spectrum from the NSO historical library https://nispdata.nso.edu/ftp/FTS_cdrom/ and read its header to get the start/stop wavenumbers, and the wavenumber spacing (DELW). I also read the data axis into an array “I” (intensities) and create an array “k” so that I can print out pairs ![]()
using FITSIO, VegaLite, DataFrames
f = FITS("881115R0.005")
# one HDU
header = read_header(f[1])
# stop/start wavenumbers
wstart=header["WSTART"]
wstop=header["WSTOP"]
# spacing
delw=header["DELW"]
# (wstop - wstart) / delw = NPO
npo=header["NPO"]
# read length of the data axis of HDU
naxis1=header["NAXIS1"]
I=read(f[1])
k=[wstart+j*delw for j in 1:npo]
# 602112-element Vector{Float32}
close(f)Some software that processes these very large spectra (this file had 602112 lines) expects the data to be stored in a binary format, which is about 20% the size of an ASCII data file, so we can print our ![]() pairs or just the
pairs or just the ![]() values to a binary file (as binary Float32 data)
values to a binary file (as binary Float32 data)
""" Write FITS file intensities to binary file"""
binwrite=function(Intensities,outfile)
open(outfile, "w") do file
for j in 1:length(Intensities)
write(file, Float32.(Intensities[j])) # write a Float64
end
endOnce we have the ![]() pairs, we can do things like find centers of gravities of lines, search for strong/high
pairs, we can do things like find centers of gravities of lines, search for strong/high ![]() lines, and make plots of parts of the spectrum. I find that the VegaLite plotting library works best for this
lines, and make plots of parts of the spectrum. I find that the VegaLite plotting library works best for this
"""COG(L,R,k,I)=Center of gravity of intensities for L<= indices <=R
k is set of wavenumbers, I is FITS intensities"""
COG=function(L,R,k,I)
sum(k[L:R,1] .* I[L:R,1])/sum(I[L:R,1])
end
"""absCOG(L,R,k,I)=Center of gravity of abs(intensities) for L<= indices <=R
k is set of wavenumbers, I is FITS intensities"""
absCOG=function(L,R,k,I)
sum(k[L:R,1] .* abs.(I[L:R,1]))/sum(abs.(I[L:R,1]))
end
"""Display(L,R,k,I) plots intensities for L<= indices <=R
k is set of wavenumbers, I is FITS intensities"""
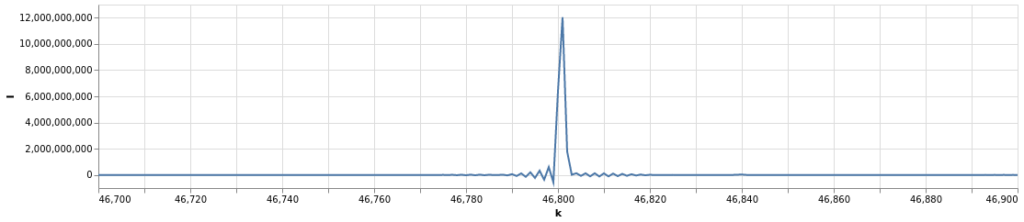
Display=function(L,R,k,I)
dat=DataFrame(k=vec(k[L:R]),I=vec(I[L:R]))
dat |> @vlplot(width=1000, :line, :k, :I)
endFor example from this datafile let’s load VegaLite and plot the region around one of the high-intensity lines using my Display function for ![]() and
and ![]() :
: